Constructing a NEP model
Introduction
In this tutorial, we will show how to use the nep executable to train a NEP potential and check the results. For background on the NEP approach see this section of the documentation.
Prerequisites
You need to have access to an Nvidia GPU with compute capability >= 3.5. CUDA 9.0 or higher is also needed to compile the GPUMD package. You can download GPUMD, unpack it, go to the src directory, and type make -j to compile. Then, you should get two executables: gpumd and nep in the src directory.
Train a NEP potential for PbTe
To train a NEP potential, one must prepare at least two input files: train.xyz and nep.in. The train.xyz file contains all the training data, and the nep.in file contains some controlling parameters. One can also optionally provide a test.xyz file that contains all the testing data.
Prepare the train.xyz and test.xyz files
The
train.xyz/test.xyzfile should be prepared according to the documentation.For the example in this tutorial, we have already prepared the
train.xyzandtest.xyzfiles in the current folder, which contain 25 and 300 structures, respectively.In this example, there are only energy and force training data. In general, one can also have virial training data.
We used the VASP code to calculate the training data, but
nepdoes not care about this. You can generate the data intrain.xyz/test.xyzusing any method that works for you. For example, before doing new DFT calculations, you can first check if there are training data publicly available and then try to convert them into the format as required bytrain.xyz/test.xyz. There are quite a few tools (https://github.com/brucefan1983/GPUMD/tree/master/tools) available for this purpose.
Prepare the nep.in file.
First, study the documentation about this input file.
For the example in this tutorial, we have prepared the
nep.infile in the current folder.The
nep.infile reads:
type 2 Te Pb
This is the simplest nep.in file, with only one keyword, which means that there are nep.txt. Therefore, the user does not need to remember the order of the atom types here. All the other parameters will have good default values (please check the screen output).
Run the nep executable
Go to the current folder from command line and type
path/to/nep # linux
path\to\nep # Windows
It took about 12 minutes to run 10,000 generations using my laptop with a GeForce RTX 2070 GPU card. One can kill the executable at any time and also restart it by simply running the executable again.
Check the training results
After running the
nepexecutable, there will be some output on the screen. We encourage the user to read the screen output carefully. It can help to understand the calculation flow ofnep.Some files will be generated in the current folder and will be updated every 100 generations (some will be updated every 1000 generations). Therefore, one can check the results even before finishing the maximum number of generations as set in
nep.in.If the user has not studied the documentation of the output files generated by
nep, it is time to read about those here.We will use Python to visualize the results in some of the output files next. We first load
pylabthat we need.
[1]:
from pylab import *
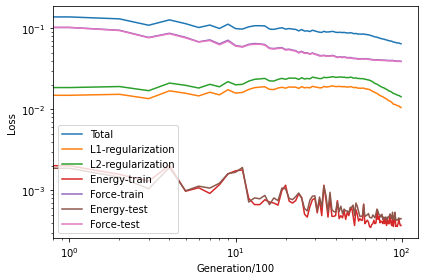
Checking the loss.out file
We see that the
The energy loss is the root mean square error (RMSE) of energy per atom, which converges to about 0.4 meV/atom.
The force loss is the RMSE of force components, which converges to about 40 meV/Å.
[2]:
loss = loadtxt('loss.out')
loglog(loss[:, 1:6])
loglog(loss[:, 7:9])
xlabel('Generation/100')
ylabel('Loss')
legend(['Total', 'L1-regularization', 'L2-regularization', 'Energy-train', 'Force-train', 'Energy-test', 'Force-test'])
tight_layout()
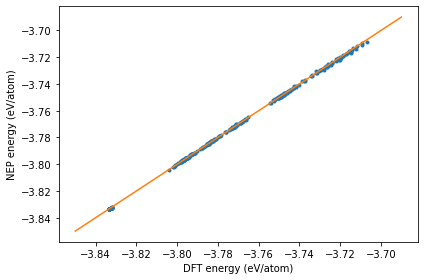
Checking the energy_test.out file
The dots are the raw data and the line represents the identity function used to guide the eyes.
[3]:
energy_test = loadtxt('energy_test.out')
plot(energy_test[:, 1], energy_test[:, 0], '.')
plot(linspace(-3.85,-3.69), linspace(-3.85,-3.69), '-')
xlabel('DFT energy (eV/atom)')
ylabel('NEP energy (eV/atom)')
tight_layout()
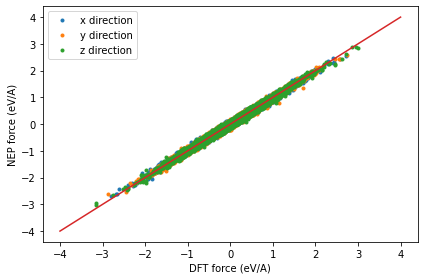
Checking the force_test.out file
The dots are the raw data and the line represents the identity function used to guide the eyes.
[5]:
force_test = loadtxt('force_test.out')
plot(force_test[:, 3:6], force_test[:, 0:3], '.')
plot(linspace(-4,4), linspace(-4,4), '-')
xlabel('DFT force (eV/A)')
ylabel('NEP force (eV/A)')
legend(['x direction', 'y direction', 'z direction'])
tight_layout()
Checking the virial_test.out file
In general, one can similarly check the virial_test.out file, but in this particular example, virial data were not used in the training process (no virial data exist in train.xyz/test.xyz).
Checking the other files
One can similarly check the other files: energy_train.out, force_train.out, and virial_train.out.
Restart
After each 100 steps, the
nep.restartfile will be updated.If
nep.restartexists, it means you want to continue the previous training. Remember not to change the parameters related to the descriptor and the number of neurons. Otherwise the restarting behavior is undefined.If you want to train from scratch, you need to delete
nep.restartfirst (better to first make a copy of all the results from the previous training).